Overview
The general workflow of long-read whole genome sequence analysis can usually includes the following steps
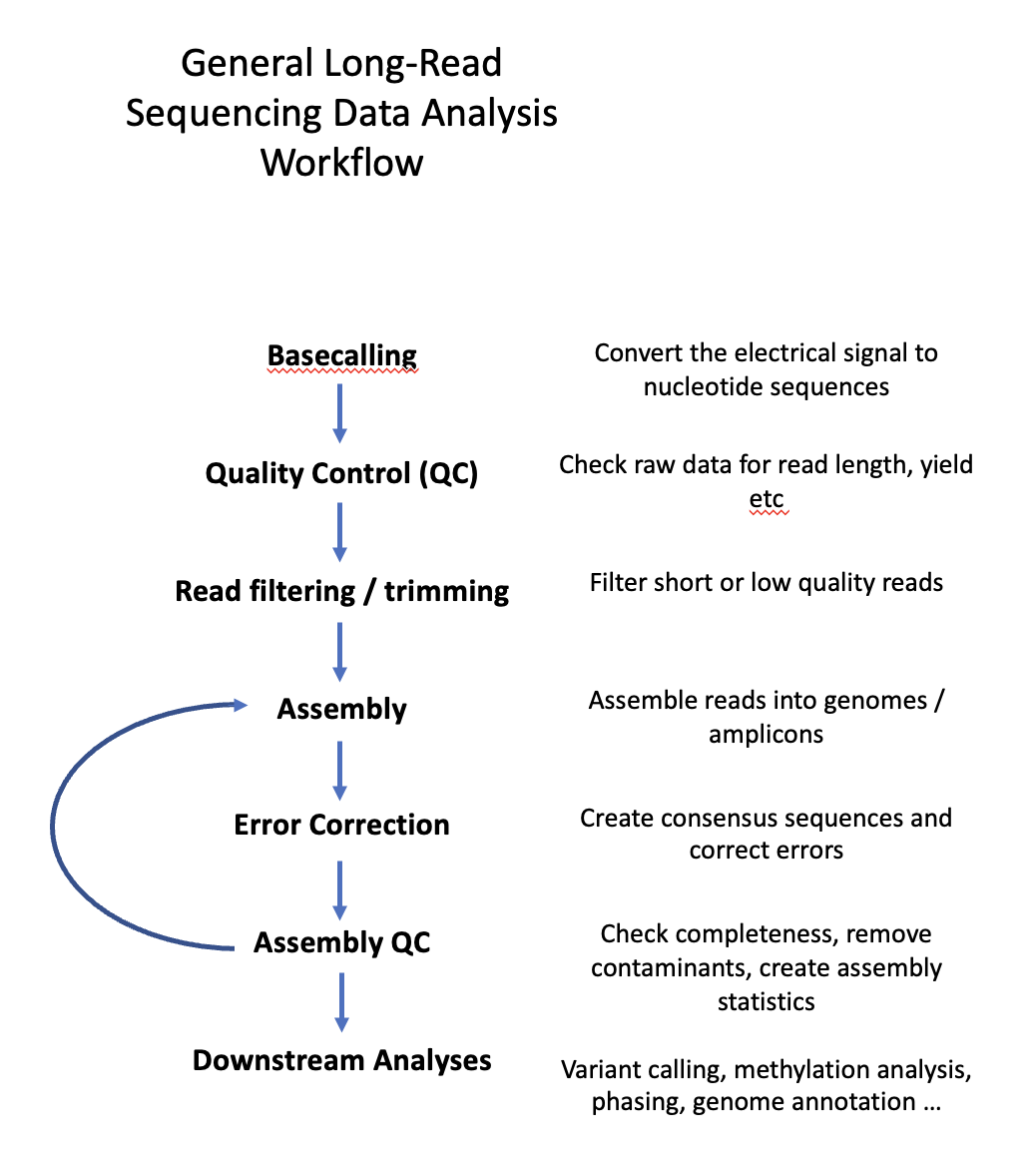
Especially the assembly step itself is often iterative depending on the input data, species sequenced and data characteristics. For example, different assemblers perform differently based on the species sequenced. Similarly, chosen cut-oofs for the QC step can affect downstream analyses. Therefore, it is common to use different assemblers and compare the outcome to achieve the best possible result.
During this course you will
- Perform basic QC
- Assemble genomes using 2 different assemblers
- Perform basic error correction
- Compare the assembly to a reference
- Perform a completeness analysis
- Perform a basic analysis of metabarcoding reads